Biopython citation information
Home » Trend » Biopython citation informationYour Biopython citation images are ready in this website. Biopython citation are a topic that is being searched for and liked by netizens today. You can Download the Biopython citation files here. Get all royalty-free images.
If you’re searching for biopython citation pictures information related to the biopython citation interest, you have pay a visit to the right site. Our website frequently gives you hints for seeing the highest quality video and image content, please kindly surf and find more informative video articles and images that fit your interests.
Biopython Citation. I have written this so far: Import csv from bio import entrez import bs4 entrez.email = my_email csvfile = open (�srdata.csv�) filereader = csv.reader (csvfile) data = list (filereader) with open (�blank.csv�,�w�) as f1: Proc natl acad sci u s a 109(12): The biopython project is a mature open source international collaboration of volunteer developers, providing python libraries for a wide range of bioinformatics problems.
 Example output of the python script list of From researchgate.net
Example output of the python script list of From researchgate.net
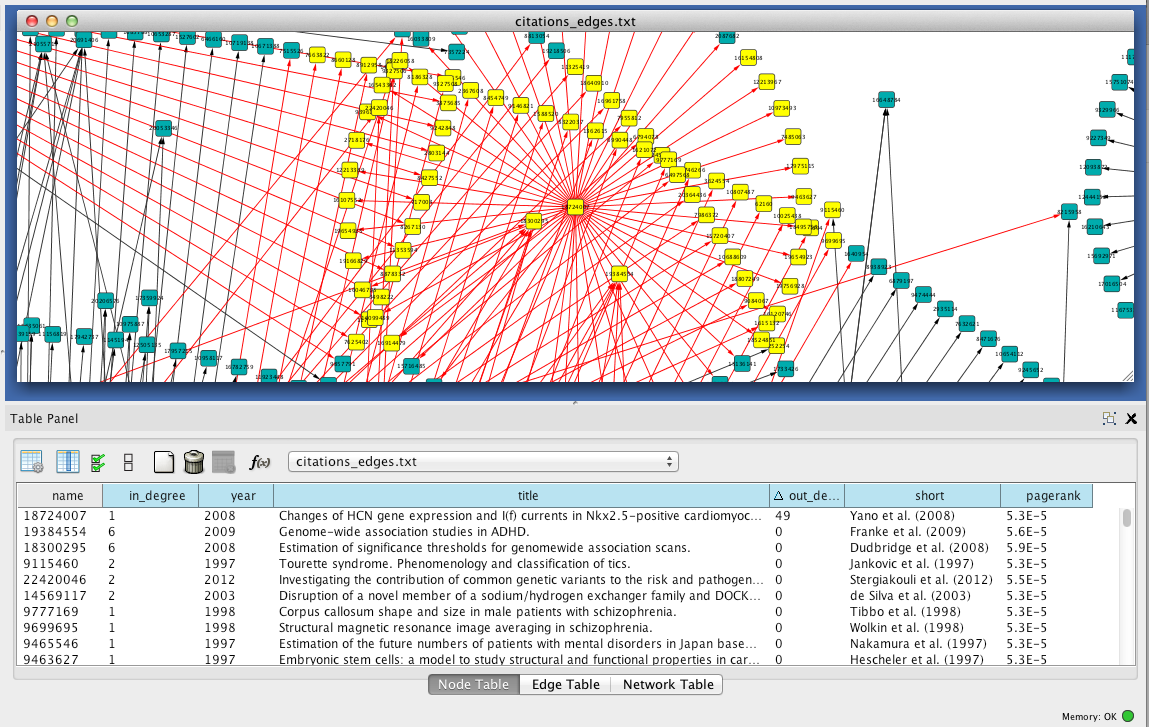
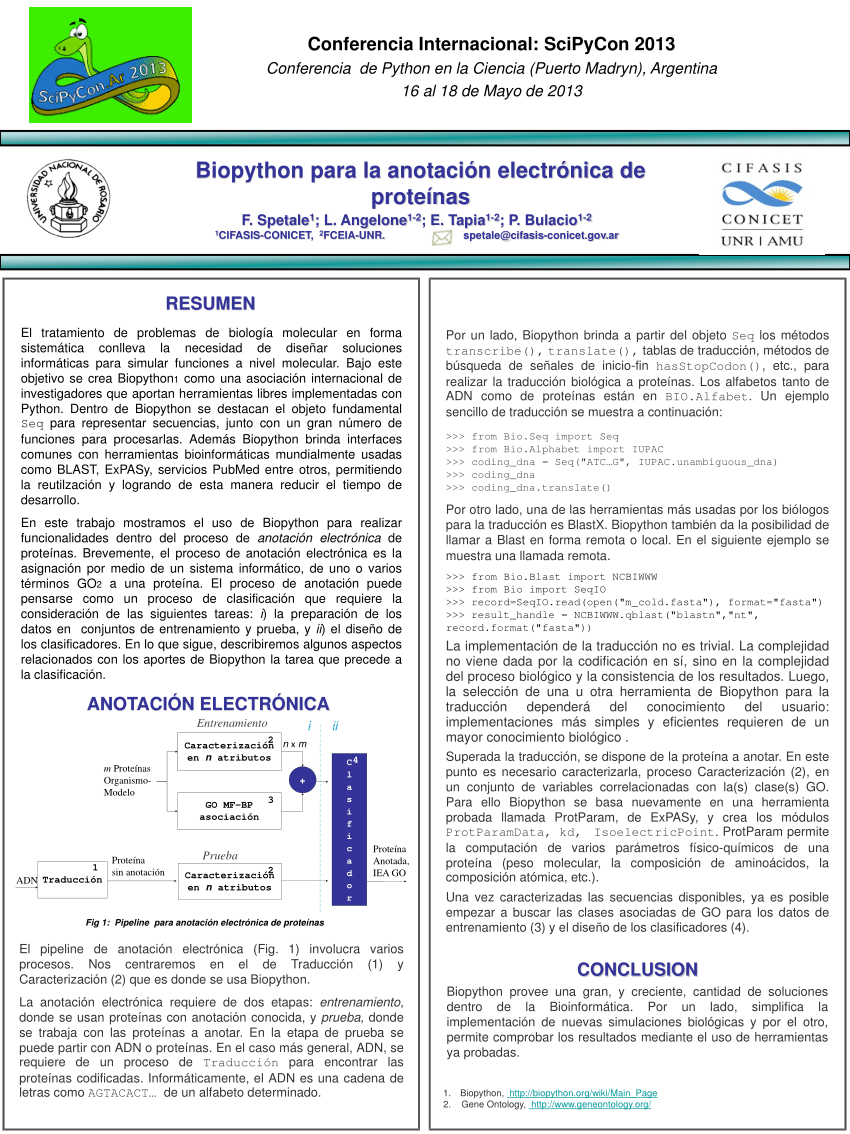
In this talk we present the current status of the biopython project (www.biopython.org), focusing on features developed in the last year, and future plans. Biopython includes modules for reading and writing different sequence file formats and multiple sequence alignments, dealing with 3d macro molecular structures, interacting with common tools such. Users can freely search for biomedical references. In this method, a list of disease eponyms is first manually collected in an excel file. Biopython and manual searching had similar numbers of citations identified with a difference per root term ranging from 0.0 to 5.56%. •biopython has modules that can directly access databases over the internet •the entrez module uses the ncbi efetch service •efetch works on many ncbi databases including protein and pubmed literature citations •the ‘gb’ data type contains much more annotation information, but rettype=‘fasta’ also works
Users can freely search for biomedical references.
Our web site lists over 100 publications using or citing biopython. Overall, the difference in citations identified was 0.1%. , 2002 ), biojava (holland et al. Freely available python tools for computational molecular biology and bioinformatics}, author= {cock, peter ja and antao, tiago and chang, jeffrey t and chapman, brad a and cox, cymon j and dalke, andrew and friedberg, iddo and hamelryck, thomas and kauff, frank and. This post describes how you can programmatically search the pubmed database with python, in order… Biopython and manual searching had similar numbers of citations identified with a difference per root term ranging from 0.0 to 5.56%.
 Source: nrnb.org
Source: nrnb.org
In this talk we present the current status of the biopython project (www.biopython.org), focusing on features developed in the last year, and future plans. Cock p and the biopython contributors. •biopython has modules that can directly access databases over the internet •the entrez module uses the ncbi efetch service •efetch works on many ncbi databases including protein and pubmed literature citations •the ‘gb’ data type contains much more annotation information, but rettype=‘fasta’ also works Bioinformatics software repository containing python scripts intended for search and download of genetic information obtained from genbank ncbi genetics data resources in support of developing pcr primers, targeted genetic databases, genetic analyses, and data interpretation. Biopython includes modules for reading and writing different sequence file formats and multiple sequence alignments, dealing with 3d macro molecular structures, interacting with common.
 Source: researchgate.net
Source: researchgate.net
I am using biopython to fill a csv file of data about citations from their pubmed title. I have written this so far: Bioinformatics software repository containing python scripts intended for search and download of genetic information obtained from genbank ncbi genetics data resources in support of developing pcr primers, targeted genetic databases, genetic analyses, and data interpretation. Since its founding in 1999 (chapman and chang, 2000), biopython has grown into a large collection of modules, described briefly below, intended for computational biology or bioinformatics programmers to use in scripts or incorporate into their own software. Biopython project update 2019 [version 1;
 Source: slideshare.net
Source: slideshare.net
I want to create a citation, so i start by trying to #get the names of the authors, but i am not sure why this is not working. As an alternative, we modified an existing biopython method for searching pubmed. Show activity on this post. I want to create a citation, so i start by trying to #get the names of the authors, but i am not sure why this is not working. , 2008 ), bioruby ( www.bioruby.org.
 Source: researchgate.net
Source: researchgate.net
This post describes how you can programmatically search the pubmed database with python, in order… Freely available python tools for computational molecular biology and bioinformatics. Since its founding in 1999 (chapman and chang, 2000), biopython has grown into a large collection of modules, described briefly below, intended for computational biology or bioinformatics programmers to use in scripts or incorporate into their own software. Includes multiple functions to streamline this process. The biopython project is a mature open source international collaboration of volunteer developers, providing python libraries for a wide range of bioinformatics problems.
 Source: researchgate.net
Source: researchgate.net
Biopython project update 2019 [version 1; Proc natl acad sci u s a 109(12): Biopython includes modules for reading and writing different sequence file formats and multiple sequence alignments, dealing with 3d macro molecular structures, interacting with common. Minor update to reflect some changes in the pubmed api pubmed is a search engine accessing millions of biomedical citations. Our web site lists over 100 publications using or citing biopython.
 Source: researchgate.net
Source: researchgate.net
Minor update to reflect some changes in the pubmed api. It is a project, which dates as far back as august 1999 [22] [23. I have written this so far: Import csv from bio import entrez import bs4 entrez.email = my_email csvfile = open (�srdata.csv�) filereader = csv.reader (csvfile) data = list (filereader) with open (�blank.csv�,�w�) as f1: , 2002 ), biojava (holland et al.
 Source: researchgate.net
Source: researchgate.net
View 2 excerpts, cites background. For some articles, the access to the full text paper is also open. Our web site lists over 100 publications using or citing biopython. I am using biopython to fill a csv file of data about citations from their pubmed title. It is important to ensure the information in square brackets after the title is included in this citation.
 Source: researchgate.net
Source: researchgate.net
Our web site lists over 100 publications using or citing biopython. Biopython project update 2019 [version 1; For some articles, the access to the full text paper is also open. Biopython includes modules for reading and writing different sequence file formats and multiple sequence alignments, dealing with 3d macro molecular structures, interacting with common. It is modeled on the highly successful bioperl project, but has the goal of making libraries available for people doing computations in python.
 Source: researchgate.net
Source: researchgate.net
Biopython and manual searching had similar numbers of citations identified with a difference per root term ranging from 0.0 to 5.56%. I have written this so far: Import csv from bio import entrez import bs4 entrez.email = my_email csvfile = open (�srdata.csv�) filereader = csv.reader (csvfile) data = list (filereader) with open (�blank.csv�,�w�) as f1: It is a project, which dates as far back as august 1999 [22] [23. Posts about biopython written by marco.
 Source: researchgate.net
Source: researchgate.net
This post describes how you can programmatically search the pubmed database with python, in order… Cock p and the biopython contributors. The oxford university press journal bioinformatics has recently published an application note describing biopython (cock et al., 2009). A python script then creates permutations of the eponyms that might exist in the cited literature. Overall, the difference in citations identified was 0.1%.
 Source: researchgate.net
Source: researchgate.net
Our web site lists over 100 publications using or citing biopython. Freely available python tools for computational molecular biology and bioinformatics. In this talk we present the current status of the biopython project (www.biopython.org), focusing on features developed in the last year, and future plans. Users can freely search for biomedical references. It is a project, which dates as far back as august 1999 [22] [23.
 Source: researchgate.net
Source: researchgate.net
Biopython includes modules for reading and writing different sequence file formats and multiple sequence alignments, dealing with 3d macro molecular structures, interacting with common tools such. •biopython has modules that can directly access databases over the internet •the entrez module uses the ncbi efetch service •efetch works on many ncbi databases including protein and pubmed literature citations •the ‘gb’ data type contains much more annotation information, but rettype=‘fasta’ also works The biopython project is a mature open source international collaboration of volunteer developers, providing python libraries for a wide range of bioinformatics problems. This post describes how you can programmatically search the pubmed database with python, in order… Biopython includes modules for reading and writing different sequence file formats and multiple sequence alignments, dealing with 3d macro molecular structures, interacting with common.
 Source: researchgate.net
Source: researchgate.net
It contains classes to represent biological sequences and sequence annotations, and it is able to read and write to a variety of file formats.it also allows for a programmatic means of accessing. Posts about biopython written by marco. Print author_name return fetch_abstract(22864638) xml looks like this: Our web site lists over 100 publications using or citing biopython. Biopython includes modules for reading and writing different sequence file formats and multiple sequence alignments, dealing with 3d macro molecular structures, interacting with common tools such.
 Source: researchgate.net
Source: researchgate.net
It is modeled on the highly successful bioperl project, but has the goal of making libraries available for people doing computations in python. The oxford university press journal bioinformatics has recently published an application note describing biopython (cock et al., 2009). Freely available python tools for computational molecular biology and bioinformatics}, author= {cock, peter ja and antao, tiago and chang, jeffrey t and chapman, brad a and cox, cymon j and dalke, andrew and friedberg, iddo and hamelryck, thomas and kauff, frank and. Biopython includes modules for reading and writing different sequence file formats and multiple sequence alignments, dealing with 3d macro molecular structures, interacting with common tools such. It is important to ensure the information in square brackets after the title is included in this citation.
 Source: researchgate.net
Source: researchgate.net
Import csv from bio import entrez import bs4 entrez.email = my_email csvfile = open (�srdata.csv�) filereader = csv.reader (csvfile) data = list (filereader) with open (�blank.csv�,�w�) as f1: The initial results show that it’s possible to use free tools to analyze data in the life sciences and consequently reduce the time and cost required to analyze acmv, and the adoption of such open source libraries in biopython is proposed. Posts about biopython written by marco. Import csv from bio import entrez import bs4 entrez.email = my_email csvfile = open (�srdata.csv�) filereader = csv.reader (csvfile) data = list (filereader) with open (�blank.csv�,�w�) as f1: Show activity on this post.
 Source: researchgate.net
Source: researchgate.net
Biopython and manual searching had similar numbers of citations identified with a difference per root term ranging from 0.0 to 5.56%. In this method, a list of disease eponyms is first manually collected in an excel file. , 2002 ), biojava (holland et al. Minor update to reflect some changes in the pubmed api pubmed is a search engine accessing millions of biomedical citations. Posts about biopython written by marco.
 Source: researchgate.net
Source: researchgate.net
Cock p and the biopython contributors. It contains classes to represent biological sequences and sequence annotations, and it is able to read and write to a variety of file formats.it also allows for a programmatic means of accessing. The biopython project is a mature open source international collaboration of volunteer developers, providing python libraries for a wide range of bioinformatics problems. Freely available python tools for computational molecular biology and bioinformatics. For all root terms, manual searching identified additional citations.
 Source: researchgate.net
Source: researchgate.net
Biopython includes modules for reading and writing different sequence file formats and multiple sequence alignments, dealing with 3d macro molecular structures, interacting with common tools such. The initial results show that it’s possible to use free tools to analyze data in the life sciences and consequently reduce the time and cost required to analyze acmv, and the adoption of such open source libraries in biopython is proposed. The biopython project is a mature open source international collaboration of volunteer developers, providing python libraries for a wide range of bioinformatics problems. Biopython includes modules for reading and writing different sequence file formats and multiple sequence alignments, dealing with 3d macro molecular structures, interacting with common tools such. In this talk we present the current status of the biopython project (www.biopython.org), focusing on features developed in the last year, and future plans.
This site is an open community for users to do submittion their favorite wallpapers on the internet, all images or pictures in this website are for personal wallpaper use only, it is stricly prohibited to use this wallpaper for commercial purposes, if you are the author and find this image is shared without your permission, please kindly raise a DMCA report to Us.
If you find this site good, please support us by sharing this posts to your favorite social media accounts like Facebook, Instagram and so on or you can also bookmark this blog page with the title biopython citation by using Ctrl + D for devices a laptop with a Windows operating system or Command + D for laptops with an Apple operating system. If you use a smartphone, you can also use the drawer menu of the browser you are using. Whether it’s a Windows, Mac, iOS or Android operating system, you will still be able to bookmark this website.
Category
Related By Category
- Apa book citation more than one author information
- Adjust citation style refworks information
- Apa citation generator online website information
- Bibtex citations information
- Apa citation format generator citation machine information
- Asa citation format information
- Apa citation format conference paper information
- Brain citation style information
- Appa citaat information
- Apa format citation online information