Bowtie2 citation information
Home » Trending » Bowtie2 citation informationYour Bowtie2 citation images are ready. Bowtie2 citation are a topic that is being searched for and liked by netizens now. You can Get the Bowtie2 citation files here. Download all free vectors.
If you’re looking for bowtie2 citation pictures information related to the bowtie2 citation interest, you have come to the ideal site. Our site always provides you with hints for refferencing the highest quality video and picture content, please kindly search and find more enlightening video articles and graphics that fit your interests.
Bowtie2 Citation. If playback doesn�t begin shortly, try restarting your device. This package provides an r wrapper of the popular bowtie2 sequencing reads aligner and adapterremoval, a convenient tool for rapid adapter trimming, identification, and read merging. Alignment comparison using hiseq 2000, 454 and ion torrent reads. The package also allows for the creation of.bam files via rsamtools.
 Coverage comparison between Bowtie2, Hisat2 and Subread From researchgate.net
Coverage comparison between Bowtie2, Hisat2 and Subread From researchgate.net
Build bowtie2 index from reference sequences. If playback doesn�t begin shortly, try restarting your device. For the human genome, its. It is particularly good at aligning reads of about 50 up to 100s or 1,000s of characters, and particularly good at aligning to relatively long (e.g. Langmead b, trapnell c, pop m, salzberg sl. Erysiphe necator, more commonly known as powdery
Please note that indel alignments, local alignments and discordant alignments are disallowed when rsem uses bowtie 2 since rsem currently cannot handle them.
The package contains wrapper functions that allow for genome indexing and alignment to those indexes. (bowtie/bowtie2gp are the fastest of the five tools tried. For the human genome, its. Bowtie2 (g p) and the original bowtie2 release were compared on bioplanet�s gcat synthetic benchmarks. It may be true that bowtie2 is the most widely used aligner, but not probable. It is especially good for aligning reads.
 Source: researchgate.net
Source: researchgate.net
These files together constitute the index: For the human genome, its. It is particularly good at aligning reads of about 50 up to 100s or 1,000s of characters, and particularly good at aligning to relatively long (e.g. Bowtie2 is a tool for fast and sensitive sequencing read alignment. This paper compares five aligners (including bowtie2, bwa, and novoalign) on several metrics such as proper pairing, speed, and sensitivity:
 Source: researchgate.net
Source: researchgate.net
Whose lengths are from 50 to hundreds or thousands of characters. The package contains wrapper functions that allow for genome indexing and alignment to those indexes. (bowtie/bowtie2gp are the fastest of the five tools tried. Build bowtie2 index from reference sequences. Alignment comparison using hiseq 2000, 454 and ion torrent reads.
 Source: researchgate.net
Source: researchgate.net
Langmead b, trapnell c, pop m, salzberg sl. If playback doesn�t begin shortly, try restarting your device. Each aligner was run using 48 geographically distinct samples of. Bowtie2 is a tool for fast and sensitive sequencing read alignment. It is particularly good at aligning reads of about 50 up to 100s or 1,000s of characters, and particularly good at aligning to relatively long (e.g.
 Source: researchgate.net
Source: researchgate.net
Erysiphe necator, more commonly known as powdery The package contains wrapper functions that allow for genome indexing and alignment to those indexes. It may be true that bowtie2 is the most widely used aligner, but not probable. Build bowtie2 index from reference sequences. Blast is by the far slowest, data not shown.) on the cancer institute’s paired end dna sequence data bowtie2gp is 26% faster than bowtie2 from which it was derived.
 Source: researchgate.net
Source: researchgate.net
If playback doesn�t begin shortly, try restarting your device. Featuredata[sequence] reference sequences used to build bowtie2 index. Download citation | which is faster: Erysiphe necator, more commonly known as powdery Whose lengths are from 50 to hundreds or thousands of characters.
 Source: researchgate.net
Source: researchgate.net
They are all that is needed to align reads to that. The package contains wrapper functions that allow for genome indexing and alignment to those indexes. Build bowtie2 index from reference sequences. Featuredata[sequence] reference sequences used to build bowtie2 index. Previously you could only select files within a folder, so this is a huge improvement in the usability of such a key new feature of galaxy ( pull request 12310 ).
 Source: researchgate.net
Source: researchgate.net
If playback doesn�t begin shortly, try restarting your device. Download citation | which is faster: Bowtie2gp bowtie > bowtie2 > bwa | we have recently used genetic programming to automatically generate an improved version of. This paper compares five aligners (including bowtie2, bwa, and novoalign) on several metrics such as proper pairing, speed, and sensitivity: The package contains wrapper functions that allow for genome indexing and alignment to those indexes.
 Source: researchgate.net
Source: researchgate.net
It is particularly good at aligning reads of about 50 up to 100s or 1,000s of characters, and particularly good at aligning to relatively long (e.g. Previously you could only select files within a folder, so this is a huge improvement in the usability of such a key new feature of galaxy ( pull request 12310 ). (bowtie/bowtie2gp are the fastest of the five tools tried. If playback doesn�t begin shortly, try restarting your device. For the human genome, its.
 Source: researchgate.net
Source: researchgate.net
This package provides an r wrapper of the popular bowtie2 sequencing reads aligner and adapterremoval, a convenient tool for rapid adapter trimming, identification, and read merging. Build bowtie2 index from reference sequences. “new insights from uncultivated genomes of the global human gut microbiome.” nature 568.7753 (2019): For the human genome, its. If playback doesn�t begin shortly, try restarting your device.
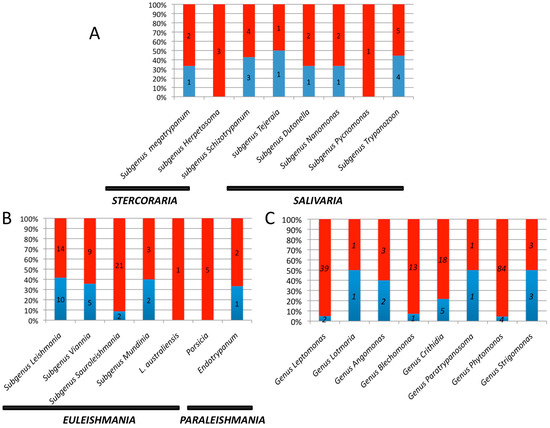 Source: mdpi.com
Source: mdpi.com
In the case of a large index these suffixes will have a bt2l termination. Build bowtie2 index from reference sequences. This paper compares five aligners (including bowtie2, bwa, and novoalign) on several metrics such as proper pairing, speed, and sensitivity: Blast is by the far slowest, data not shown.) on the cancer institute’s paired end dna sequence data bowtie2gp is 26% faster than bowtie2 from which it was derived. It is particularly good at aligning reads of about 50 up to 100s or 1,000s of characters, and particularly good at aligning to relatively long (e.g.
 Source: researchgate.net
Source: researchgate.net
Whose lengths are from 50 to hundreds or thousands of characters. Bowtie2 (g p) and the original bowtie2 release were compared on bioplanet�s gcat synthetic benchmarks. Please note that indel alignments, local alignments and discordant alignments are disallowed when rsem uses bowtie 2 since rsem currently cannot handle them. The package also allows for the creation of.bam files via rsamtools. Blast is by the far slowest, data not shown.) on the cancer institute’s paired end dna sequence data bowtie2gp is 26% faster than bowtie2 from which it was derived.
 Source: researchgate.net
Source: researchgate.net
Langmead b, trapnell c, pop m, salzberg sl. Download citation | which is faster: It may be true that bowtie2 is the most widely used aligner, but not probable. Bowtie2 · bowtie2 to give bowtie2gp, we have recovered the lost speed and retained the additional functionality. Please note that indel alignments, local alignments and discordant alignments are disallowed when rsem uses bowtie 2 since rsem currently cannot handle them.
 Source: researchgate.net
Source: researchgate.net
Bowtie2 (g p) and the original bowtie2 release were compared on bioplanet�s gcat synthetic benchmarks. The package contains wrapper functions that allow for genome indexing and alignment to those indexes. Please note that indel alignments, local alignments and discordant alignments are disallowed when rsem uses bowtie 2 since rsem currently cannot handle them. Download citation | which is faster: Blast is by the far slowest, data not shown.) on the cancer institute’s paired end dna sequence data bowtie2gp is 26% faster than bowtie2 from which it was derived.
 Source: researchgate.net
Source: researchgate.net
These files together constitute the index: Add additional flags for each option. For the human genome, its. This package provides an r wrapper of the popular bowtie2 sequencing reads aligner and adapterremoval, a convenient tool for rapid adapter trimming, identification, and read merging. The package contains wrapper functions that allow for genome indexing and alignment to those indexes.
 Source: dl.acm.org
Source: dl.acm.org
In the case of a large index these suffixes will have a bt2l termination. Article evaluation and assessment of. Bowtie2 (g p) enhancements were also applied to the latest bowtie2 release (2.2.3, 29 may 2014) and retained both the gp and the manually introduced improvements. For the human genome, its. (bowtie/bowtie2gp are the fastest of the five tools tried.
Source: researchgate.net
Whose lengths are from 50 to hundreds or thousands of characters. Build bowtie2 index from reference sequences. This package provides an r wrapper of the popular bowtie2 sequencing reads aligner and adapterremoval, a convenient tool for rapid adapter trimming, identification, and read merging. Previously you could only select files within a folder, so this is a huge improvement in the usability of such a key new feature of galaxy ( pull request 12310 ). Explore citation contexts and check if this article has been supported or.
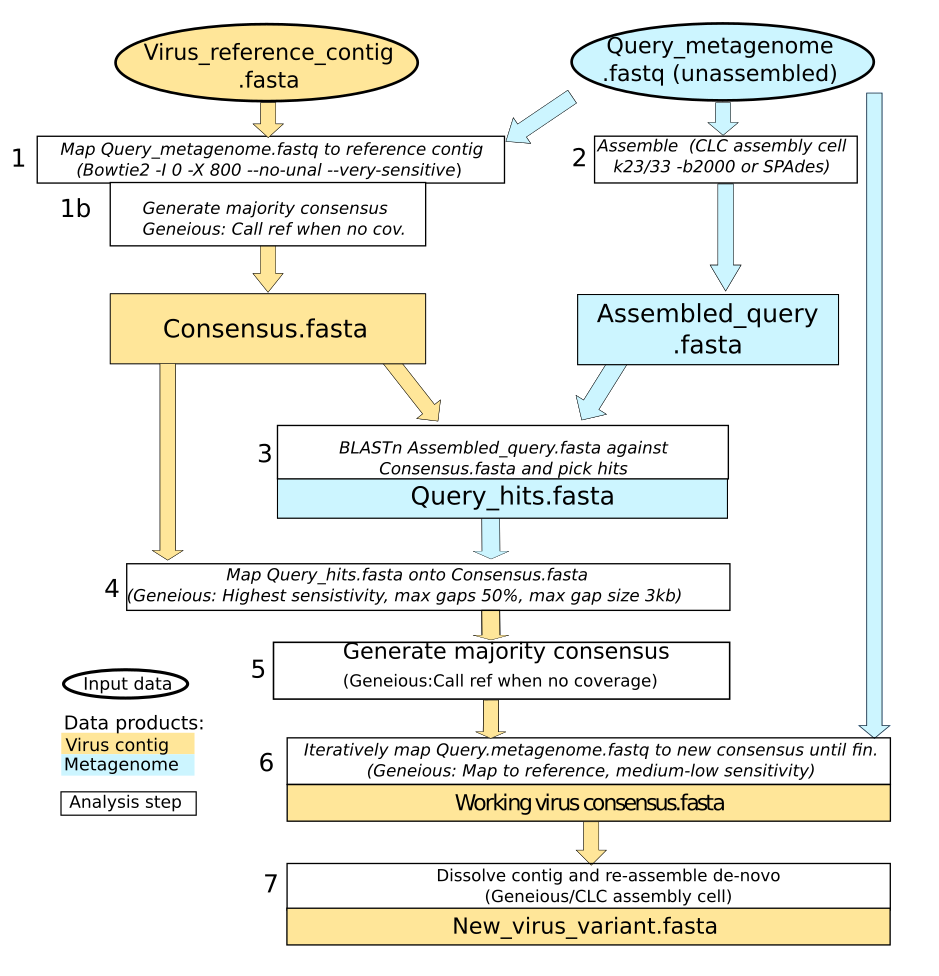 Source: protocols.io
Source: protocols.io
Build bowtie2 index from reference sequences. The package also allows for the creation of.bam files via rsamtools. This package provides an r wrapper of the popular bowtie2 sequencing reads aligner and adapterremoval, a convenient tool for rapid adapter trimming, identification, and read merging. Erysiphe necator, more commonly known as powdery Blast is by the far slowest, data not shown.) on the cancer institute’s paired end dna sequence data bowtie2gp is 26% faster than bowtie2 from which it was derived.
 Source: manual.omicsbox.biobam.com
Source: manual.omicsbox.biobam.com
Build bowtie2 index from reference sequences. Bowtie2 (g p) and the original bowtie2 release were compared on bioplanet�s gcat synthetic benchmarks. Each aligner was run using 48 geographically distinct samples of. Build bowtie2 index from reference sequences. The package contains wrapper functions that allow for genome indexing and alignment to those indexes.
This site is an open community for users to do sharing their favorite wallpapers on the internet, all images or pictures in this website are for personal wallpaper use only, it is stricly prohibited to use this wallpaper for commercial purposes, if you are the author and find this image is shared without your permission, please kindly raise a DMCA report to Us.
If you find this site convienient, please support us by sharing this posts to your preference social media accounts like Facebook, Instagram and so on or you can also save this blog page with the title bowtie2 citation by using Ctrl + D for devices a laptop with a Windows operating system or Command + D for laptops with an Apple operating system. If you use a smartphone, you can also use the drawer menu of the browser you are using. Whether it’s a Windows, Mac, iOS or Android operating system, you will still be able to bookmark this website.
Category
Related By Category
- Autonomie citaten information
- Apa citation maker website information
- 10 citate despre iubire information
- 2018 journal citation reports information
- Blik op de weg citaat information
- Belle citation courtes information
- Baudelaire citation information
- Apa citation sample book information
- Aya ezawa google citations information
- Apa citation generator for government website information